
Isaac Vock ‘25
(he/him)
Home Department: Molecular Biophysics and Biochemistry (Simon Lab)
Research Project: The EZbakR suite: computational tools to facilitate studies of gene expression dynamics and regulation
I am developing a software suite that will facilitate analyses of the mechanisms of gene expression using high-throughput RNA sequencing (RNA-seq). I am particularly interested in developing improved analyses of nucleotide recoding RNA analyses (NR-seq; TimeLapse-seq, SLAM-seq, TUC-seq, etc.), a collection of extensions of RNA-seq developed by the Simon lab and others to investigate the kinetics of RNA metabolism. The primary focus of my work thus far has been to develop an R package (Bayesian analysis of the kinetic of RNA; bakR) that performs statistically rigorous analyses of NR-seq methods that assess the kinetics of bulk RNA synthesis and degradation. I am currently working on developing a significant improvement and extension of bakR called EZbakR, and am using this tool to study post-transcriptional regulation of transcript isoforms.
Relevant Publications
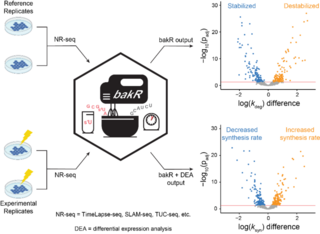
bakR is an R package implementing a novel hierarchical model to identify differences in RNA synthesis rates and stabilities using TimeLapse-seq, SLAM-seq, TUC-seq, or similar metabolic labeling methods.